Can Stress Produce Resistance to Cancer Therapies?
We all experience some level of stress in our daily lives. However, high levels of chronic stress can negatively impact the human body and can contribute to health problems, such as high blood pressure, heart disease, obesity and diabetes. Studies now show that chronic stress can affect every system in the body, including the nervous, gastrointestinal, cardiovascular, respiratory and reproductive systems.
Based on the broad effects stress can have on the body, could chronic stress contribute to cancer therapy resistance? There are multiple known mechanisms that result in higher rates of mutagenesis or an increase in the rate of specific genetic changes during stress. These mechanisms are called stress-induced mutagenesis (SIM). SIM is a key resistance mechanism of therapy due to the rapid expansion of genetic diversity. SIM-dependent identification of targets to minimize resistance to anticancer therapy could be a great strategy for treating cancer patients.
Many microorganisms including tumor cells, result in increased mutation or genetic recombination rates when exposed to a stressful environment. This can increase the probability that the population acquires adaptations that allow it to avoid extinction. Because of this, it has been suggested that the increase in the production rate of genetic variation is itself an adaptation for the survival of cells.
Case Study
In a study by Cipponi A et al, 2020 (PMID: 32499442); multiple human cancer cell lines were selected and treated with targeted therapy in a dose-response manner. The drug
concentrations used matched near-extinction conditions, but were less than lethal concentrations. The cell lines were selected and treated to develop the resistance. Resistance cells were analyzed for genetic and molecular features along with drug sensitivity. Most of the resistance cells elevated DNA Double-stranded break (DSB). This mechanism was linked with stress-induced mutagenesis.
The 94T778 cell line identified as genomically unstable, showing amplification of oncogenes CDK4 and MDM2. This was exposed to palbociclib (CDK4/6 Inhibitor) and nutlin (an inhibitor of the p53–MDM2 interaction). Surprisingly, C238F resistance mutation in TP53 was detected at 11 weeks under nutlin-3a. These data support a SIM-induced intrinsic fitness penalty early during adaptation.
The MTOR signaling pathway is ubiquitously expressed in human tissues and functions as an evolutionarily conserved sensor of environmental and endogenous stress. The 94T778, SKMEL28, and human breast cancer SKBR3 cell lines were engineered with MTOR directed shRNAs and were exposed to tunicamycin, vemurafenib, and the EGFR/HER2 inhibitor lapatinib, respectively. All cell lines showed a reduction in MTOR protein levels and a reduced phosphorylation of the downstream kinase p70-S6K. A key finding of Cipponi A et al, 2020 is inhibition of mTOR promoted adaptive mutagenesis by repressing DNA repair mechanism. MTOR silencing increased heterogenicity. In the in vivo pancreatic cell PDX model, mTOR repression accelerates Palbociclib resistance emergence due to increased heterogenicity. Conversely, pharmacological inhibition of MTOR repressed the homologous recombination and Fanconi anemia pathway but not the error prone DNA Polymerase. Cipponi A et al, 2020 explained the effect of stress induced mutagenesis and identified MTOR as a key responsible pathway but did not identify the key mechanistic pathways involved in between MTOR pathway and SIM. Cellworks provided the further insight considering the holistic approach at detailed cellular level network considering all cell signaling, epigenetic level regulation, metabolomics, transcriptomics and transcriptomic level integrated network.
Cellworks Insight
We used a novel predictive dynamic simulation network to look into the role of MTOR pathway in mutagenesis and identified the various biological network by which this effect could be possible. The predictive simulation technology comprehensively incorporates integrated networks of signaling and metabolic pathways that underlie all cancer phenotypes. The predictive simulation approach differs from other pathway modeling approaches. Unlike a static network, big data mining or informatics approach, this approach enables novel drug simulations and predictions prospectively validated using clinical and biomarker endpoints.
The integrated cancer physiology network that covers all disease phenotypes in the simulation model are aggregated through manual scientific review one reaction at a time to maintain a high quality of input information and address issues of prevalent contradictory datasets. In addition, the network is continuously enhanced with information from new research. This predictive platform was then used to test a library of molecularly targeted drugs on the patient simulation avatar.
Cellworks network-based analysis was able to give more insight into the connection between mTOR and DNA repair pathway which could further address the reason why MTOR silencing increased heterogenicity due to increased mutagenesis which was consequence of DNA damage.
Scientific Rationale
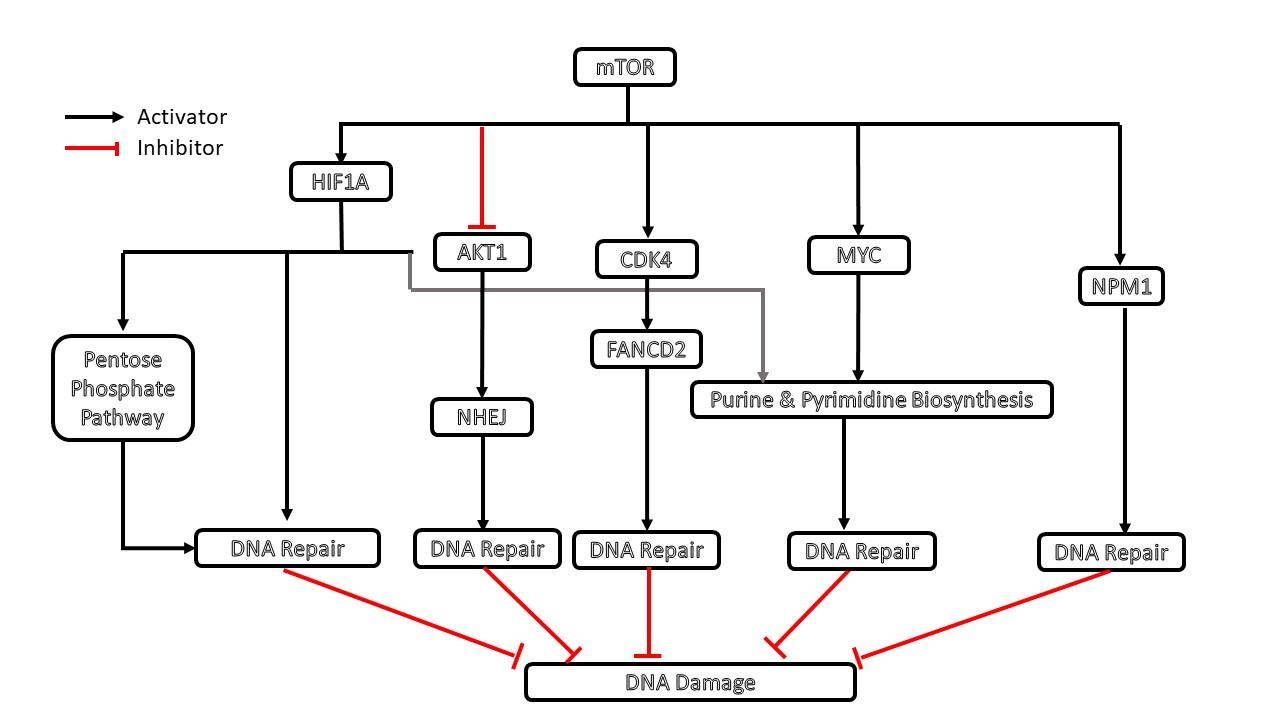
Cellworks biological-derived network reveals the connection between mTOR pathway and various repair pathway. mTOR activates HIF1A which is key transcription factor of genes like XPC and XPA which regulates FA Core complex during DNA repair. MTOR silencing could lead to impaired DNA repair and increased DNA Damage. Similarly, HIF1A is a key transcription factor of various pentose phosphate pathway enzymes. mTOR silencing mediated reduction of HIF1A results in reduced Pentose phosphate pathway. Ribose 5 phosphate is a key product of this pathway and this is required for the backbone of DNA. Reduced Ribose 5 phosphate increases damaged DNA.
MTOR is a key translation regulator of proliferative kinase CDK4 which further activates FANCD2, a one of the key enzymes of Fanconi Anemia pathway of DNA repair. MTOR silencing mediated reduced Fanconi Anemia repair pathway leads to increased DNA damage. mTOR also activates NPM1, which is one of the key regulators of MMR (Mis Match Repair) pathway. MTOR mediated silencing also results in increased DNA via reduced NPM1 level.
MTOR is also a key regulator of MYC which transcribes various enzymes of purine and pyrimidine pathway. MTOR silencing mediated reduction of MYC transcription factor reduces purine and pyrimidine biosynthesis which further reduced repair pathway and increased damage DNA.
In Conclusion
Chronic stress can affect every system in the body, including the nervous, gastrointestinal, cardiovascular, respiratory and reproductive systems. Stress-induced mutagenesis (SIM) are the genetic changes that occur due to chronic stress. SIM is a key resistance mechanism of therapy due to rapid expansion of genetic diversity. This can increase the probability that the population acquires adaptations that allow it to avoid extinction. Because of this, it has been suggested that the increase in production rate of genetic variation is itself an adaptation for survival of cells.
Cipponi A et al used drug concentrations required for near-extinction conditions, which were less than lethal concentrations, to develop cancer resistance. Resistance cells were analyzed for genetic and molecular features along with drug sensitivity. Cipponi A et al, explained the effect of stress induced mutagenesis and identified MTOR as a key responsible pathway but did not identify the key mechanistic pathways involved in between MTOR pathway and SIM.
Cellworks used a novel predictive dynamic simulation network to look into the role of MTOR pathway in mutagenesis and identified the various biological network by which this effect could be possible. Cellworks predictive simulation demonstrated a connection between mTOR and DNA repair pathway and supports the findings of why MTOR silencing increased heterogenicity due to increased mutagenesis, which was a consequence of DNA damage. The Cellworks network-based analysis showed various pathways and links, which supported the key finding of Cipponi A et al, 2020 (PMID: 32499442).




